Rearranging the tree of life: a closer look at Ctenophores
By Shannon Moorhead, RJD Intern
At first glance, members of the animal phylum Ctenophora don’t look like much. Commonly known as the comb jellies, ctenophores vaguely resemble true jellies of the phylum Cnidaria: marine organisms with translucent, gelatinous bodies that spend the majority of their time suspended in the water column. Like cnidarians, comb jellies utilize tentacles with specialized cells to capture prey, usually zooplankton and animal larvae. However, instead of nematocysts, stinging cells typical of jellyfish and anemones, to subdue prey, ctenophores have developed colloblasts, cells found in no other metazoan, animal, group (Ryan et al. 2013). These colloblasts produce a glue-like substance; the animal extends its tentacles, colloblasts adhere to plankton passing by, and then the tentacles are retracted and food particles are moved to the mouth (Moroz et al. 2014). Ctenophores also have a relatively uncommon form of locomotion for metazoans of their size. Comb jellies propel themselves by beating hair-like cilia, arranged into eight tracts called comb rows, in unison. The comb rows run the length of the animal, from the aboral to oral surface (side of organism without and with mouth, respectively) and their beating is controlled by the aboral organ, a primitive brain consisting of sensory apparatuses for gravity, balance, and light (Moroz 2015). This is one major component of a fairly complex nervous system that also includes two nerve nets, nerves for controlling the tentacles, and basic mechano- and chemoreceptors, sensory cells that take in physical and chemical information, respectively, from the surrounding environment (Moroz 2015). With such complicated methods of hunting and locomotion, it is no surprise that ctenophores need an extensive nervous system to effectively coordinate various parts of the body.
The fragile nature of ctenophores makes them hard to work with and, coupled with their lack of fossil record and available genomic data, they have been difficult to place on the phylogenetic tree of life. Trees based on a variety of aspects, from morphological characteristics to ribosomal RNA analysis have placed ctenophores in a plethora of groups. However, recent genomic analysis, coupled with analysis of the nervous system, supports a new location of ctenophores on the tree of life: as the sister group to all other Metazoan. This would imply that comb jellies were the first group of animals to branch off on its own, and may mean that they are the closest living possible representation of the ancestral metazoan all animal life evolved from.
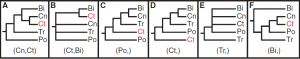
Previously proposed phylogenetic groupings of major metazoan lineages. Ct, Ctenophora; Po, Porifera; Tr, Placozoa; Cn, Cnidaria; Bi, Bilateria (Ryan et al. 2013)
When thoroughly examined, stark differences between the genome, nervous system, and development of ctenophores and other metazoan groups (particularly those with nervous systems, bilateria and Cnidaria) can be found. In their 2013 study, Ryan et al. sequenced the genome of the ctenophore Mnemiopsis leidyi, and determined that gene content data substantially supported the position of Ctenophora as a sister group to other animals. Genomic analysis of the ctenophore Pleurobrachia bachei by Moroz et al. in 2014 had similar findings. Notably, several genes known to contribute to development and patterning, the processes by which undifferentiated cells become specific structures during the growth of an organism from zygote to larva to adult, in other metazoans are completely absent. For example, HOX genes, which are essential for controlling the body plan of bilaterians during development, are missing from ctenophores (Moroz 2015). Not surprisingly, Moroz et al. 2014 found that Pleurobrachia express a multitude of genes not present in other metazoans early on in their development, suggesting ctenophores may have evolved an entirely unique set of development and patterning genes.
Comb jellies also have a variety of unique innovations, as well as the absence of usually essential metazoan genes, that indicate the independent evolution of their nervous system. Moroz 2015 found the neurons of ctenophores had a non-polarized morphology: any part of the neuron can form a synapse onto another cell, far different from the typical bilaterian neuron with its signal-receiving dendrites and signal-sending axon. Neuronal cell structures also form a “presynaptic triad”, a way of organizing the area of the neuron that sends transmits the signal to another cell that is unique to ctenophores (Moroz 2015). Microscopy reveals a row of diverse, similarly-sized vesicles containing neurotransmitters just under the cell membrane, nearest to the synapse. Behind the row of vesicles is a layer of smooth endoplasmic reticulum, followed by one or several large mitochondria.
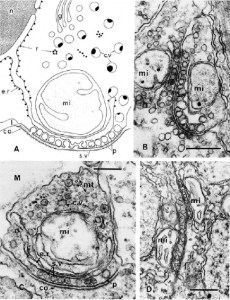
(A) drawing of an asymmetrical synapse with presynaptic triad. (B) Two-way synapse, with opposing presynaptic triads. (C) Asymmetrical synapse. (D) Soma-soma reciprocal synapse. c.v, cytoplasmic vesicles; e.r., endoplasmic reticulum; g, Golgi complex; M, mesoglea; mi, mitochondrion; mt, microtubules; r, ribosomes; s.v., synaptic vesicle (Moroz 2015).
Observations of ctenophores via microscope also show they have a high diversity of synapses compared to other metazoa, though the most prevalent is the asymmetrical synapse. The asymmetrical synapse is polarized, like in bilaterians, with one cell sending and one receiving a signal; the abundance of this type of synapse indicates that comb jellies primarily use unidirectional chemical transmitters to send a message from a neuron to another neuron, or another kind of cell (Moroz 2015). Ctenophores also utilize soma-soma reciprocal synapses, where cells synapse onto each other at staggered locations, and two-way synapses, which are not found in vertebrates. These two-way synapses involve the presynaptic triads of two neurons facing each other, meaning that signals are being transmitted in both directions across the synapse (Moroz 2015). In addition to chemical synapses, ctenophores have gap junctions, electrical synapses formed by a protein connecting the cytoplasms of adjacent cells. Moroz 2015 found that Pleurobrachia make twelve different gap junction proteins, though not connexin, the one most commonly used in chordates. These gap junctions are most common in the aboral organ, tentacles, and combs, where the quick signal transmission allows for better coordination of movements.
Evidence suggests ctenophores have adapted their own set of neurotransmitters as well. Moroz et al. 2014 was unable to detect common metazoan neurotransmitters such as acetylcholine, dopamine, serotonin, noradrenaline, adrenaline, and nitric oxide in Pleurobrachia. They also found no genes encoding enzymes to make, as well as no genes encoding receptors for, these transmitters. In the study, Moroz et al. determined that L-glutamate, an amino acid, is the most likely candidate for a ctenophore neurotransmitter. This is supported by the ability of L-glutamate to cause muscle contractions along with the unprecedented variety of ionotropic glutamate receptors (iGluRs) in comb jellies. During ctenophore development, iGluRs become present around the same time as neurons, indicating the two are associated and further supporting the candidacy of L-glutamate as a neurotransmitter (Moroz et al. 2014).
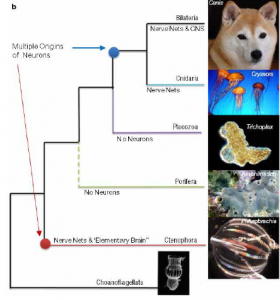
Phylogenetic tree based on recent studies, indicates the evolution of neurons at two separate points (Moroz et al. 2014)
When examined closely, the glaring differences between the genes, development, and nervous system of ctenophores and other metazoans have significant implications for the understanding of animal evolution. Ctenophores, and not sponges (Porifera) as previously thought, as the earliest diverging metazoan lineage would suggest that the nervous system evolved twice in metazoan groups, once in ctenophores and again in cnidarians and bilaterians. The members of Porifera lack a true nervous system and most likely never had one; once an animal lineage has developed a nervous system it is far too valuable to lose. This suggests that the common metazoan ancestor sponges, and also ctenophores, evolved from had not yet developed a nervous system. If this was the case, then ctenophores and cnidarians/bilaterians had to develop nervous systems completely separately, hence the unique features of the ctenophore nervous system. Convergent evolution of this complex a structure is unprecedented in the animal kingdom and this, along with other data gathered from ctenophores, could incite a complete reworking of the theories of animal evolution.
References
Moroz, L. (2015). Convergent evolution of neural systems in ctenophores. Journal of Experimental Biology, 218, 598-611.
Moroz, L. et al. (2014). The ctenophore genome and the evolutionary origins of neural systems. Nature, 000, 1-6.
Ryan, J. et al. (2013). The Genome of the Ctenophore Mnemiopsis leidyi and Its Implications for Cell Type Evolution. Science, 342, 1242592-1242592.



Leave a Reply
Want to join the discussion?Feel free to contribute!