DNA Barcoding: What is it and how can it help stranded marine mammals?
By Hannah Calich, RJD Graduate Student and Intern
Prior to 2003, when someone wanted to identify a biological specimen they would examine its morphological features (such as the shape, size, or colour of specific body parts). However, identification wasn’t possible with degraded specimens. To combat this, in 2003 Paul Hebert proposed “DNA barcoding” as a way to help identify animals without using morphological measurements (CBOL, 2014).
DNA barcoding uses small pieces of the genetic sequence obtained from a specimen’s DNA sample to determine what species the sample came from. Since the pieces all come from the same area within the DNA sequence, they can be compared to help determine the animal’s species. This concept is similar to how barcodes are used at the grocery store. To the untrained eye all barcodes look very similar, but scanners are able to identify distinct patterns in the barcode and tell the cashier what the product is. To date, over 140,000 animals, 52,000 plants, and 15,000 Fungi and other life forms have had their DNA catalogued for barcoding (CBOL, 2014).
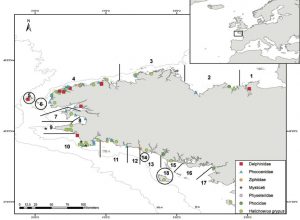
The implications of DNA barcoding extend well beyond simply creating a database. In fact, a recent study by Alfonsi et al. (2013) aimed to investigate the feasibility of using DNA barcoding to help monitor marine mammal biodiversity through strandings along the French Atlantic coast. In the last 10 years over 1,500 marine mammals from 19 different species have stranded along the French Atlantic coast (Figure 1). Unfortunately, 16.8% of the animals (258 animals) could not be identified to the species level due to body decomposition (Figure 2), poor weather conditions, or because the animal was very rare. Identifying these unknown species is important to help researchers determine what species are stranding, where they are stranding, and what (if anything) humans can do to help save these animals.
Alfonsi et al. (2013) successfully analyzed DNA samples from 92 marine mammals. The data helped confirm that animals were being correctly identified based on morphology when morphological measurements were possible. The data also helped to identify rare species and identify specimens that were too degraded or incomplete to identify based on morphology alone. In addition, Alfonsi et al. (2013) proposed that DNA barcoding could be used to monitor population movements. By combining their findings with the ongoing database, Alfonsi et al. (2013) determined that the two most commonly stranded species were the short-beaked common dolphin (Delphinus delphis) and the gray seal (Halichoerus grypus).

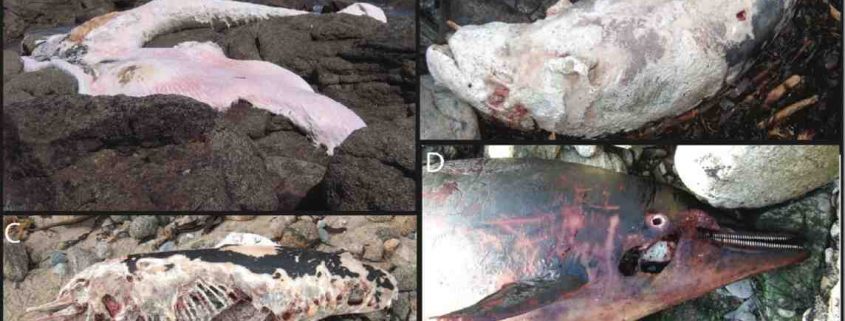
Examples of stranded marine mammals that were identified to the species level using DNA barcoding by Alfonsi et al. (2013). A-Fin Whale (Balaenoptera physalus), B-Risso’s Dolphin (Grampus griseus), C-Short-beaked common dolphin (Delphinus delphis), D-Striped dolphin (Stenella coeruleoalba)
This study was the first to demonstrate that DNA barcoding can be used to monitor marine mammal diversity through strandings data. Additionally, Alfonsi et al. (2013) showed that even when a carcass is severely degraded, good quality DNA samples can still be obtained. While more work is necessary to fine-tune identifying closely related species (e.g., within the Delphininae family), DNA barcoding has the potential to greatly increase the amount of usable data researchers can obtain from marine mammal strandings.
References:
Alfonsi E, Méheust E, Fuchs S, Carpentier F-G, Quillivic Y, Viricel A, Hassani S, Jung J-L (2013) The use of DNA barcoding to monitor the marine mammal biodiversity along the French Atlantic coast. In: Nagy ZT, Backeljau T, De Meyer M, Jordaens K (Eds) DNA barcoding: a practical tool for fundamental and applied biodiversity research. ZooKeys 365: 5–24. doi: 10.3897/zookeys.365.5873 Posterior probabilities for species identification determined by the nMDS analysis. doi: 10.3897/zookeys.365.5873.app2
CBOL (2014) What is DNA Barcoding? Received from: http://www.barcodeoflife.org/content/about/what-dna-barcoding



Leave a Reply
Want to join the discussion?Feel free to contribute!